Annotation of this kernel: https://www.kaggle.com/code/benjenkins96/understanding-survival-analysis
Understanding Survival Analysis
Explore and run machine learning code with Kaggle Notebooks | Using data from CIBMTR - Equity in post-HCT Survival Predictions
www.kaggle.com
Initial EDA
# Check the distribution of the target variables
plt.figure(figsize=(10, 5))
sns.countplot(data=train, x='efs', palette='coolwarm')
plt.title('Distribution of Event-Free Survival (efs)')
plt.show()
- Event-free survival(efs) is an important outcome measure in medical research, particularly in transplant studies
- EFS refers to the period from the start of treatment(transplant) until the occurrence of an "event": probably death in this competition
- EFS differs from Overall Survival (OS):
- OS only considers survival/death
- EFS includes not only survival but also various important clinical events related to treatment success
plt.figure(figsize=(10, 5))
sns.histplot(data=train, x='efs_time', bins=30, kde=True, color='blue')
plt.title('Distribution of Time to Event-Free Survival (efs_time)')
plt.show()
plt.figure(figsize=(6, 3))
plt.hist(train.efs_time[train.efs == 0], bins=50, label='efs=0: Patient Still Alive Or Unknown', alpha=0.5)
plt.hist(train.efs_time[train.efs == 1], bins=50, label='efs=1: Patient Dies', alpha=0.5)
plt.legend()
plt.xlabel('Event Free Survival Time')
plt.ylabel('Count')
plt.title('Histogram of Time to Event-Free Survival (efs_time)')
plt.show()
# Explore distribution of key demographic features
demo_features = ['race_group', 'sex_match', 'ethnicity']
for feature in demo_features:
plt.figure(figsize=(10, 5))
sns.countplot(data=train, x=feature, palette='viridis', order=train[feature].value_counts().index)
plt.title(f'Distribution of {feature}')
plt.xticks(rotation=45)
plt.show()
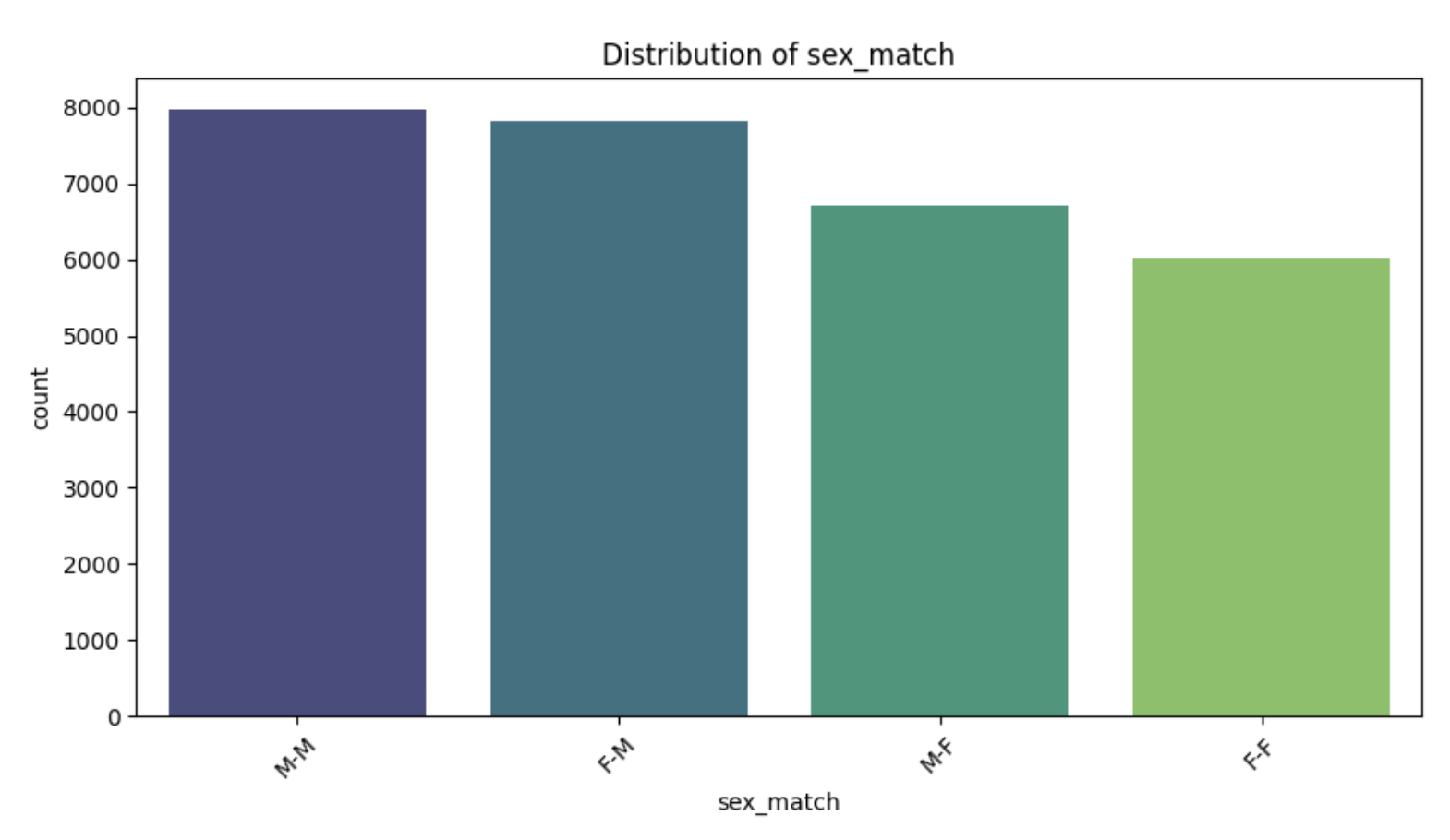

- sex_match is a variable that indicates the gender match between donor and recipient in Hematopoietic Cell Transplantation (HCT).
- It is typically categorized as follows:
- M-M: Male donor → Male recipient
- M-F: Male donor → Female recipient
- F-M: Female donor → Male recipient
- F-F: Female donor → Female recipient
Kaplan-Meier Estimator
- The Kaplan-Meier Estimator is a non-parametric statistical method used in survival analysis to estimate the survival function from time-to-event data.
- It calculates the probability that an individual will survive beyond a certain point in time, accounting for censored data (cases, where the event of interest has not occurred by the end of the study or the individual, is lost to follow-up).

Key Properties:
- The Kaplan-Meier curve is a step function, with drops occurring at times when events are observed.
- It handles censoring by only considering individuals at risk just before each event time.
Advantages:
- Non-parametric: Makes no assumptions about the distribution of survival times.
- Handles Censoring: Incorporates censored data effectively.
- Easy Interpretation: Provides intuitive survival probabilities.
Limitations:
- Assumes Independence of Censoring: Assumes that the censored individuals have the same survival prospects as those still under observation.
- Lack of Multivariable Adjustments: Does not account for the effects of covariates (e.g., age, race). For this, models like Cox regression are used.
- Uncertainty at Long Times: If few individuals remain at risk at later time points, the estimates may become less reliable.
Use Case:
In the context of HCT survival analysis:
- Kaplan-Meier can estimate survival probabilities for the entire population or subgroups (e.g., race or gender).
- It helps visualize differences in survival rates among groups, providing insights into disparities or the impact of certain factors.
Results:
- The Kaplan-Meier survival curve represents the probability of remaining event-free (e.g., alive or without relapse) over time, with the y-axis showing survival probability and the x-axis representing time in months.
- In Kaplan-Meier survival curves, "event-free (alive or without relapse)" means satisfying both of these conditions:
- alive: the patient is living
- without relapse: the disease has not recurred(재발)
- alive: the patient is living
- In Kaplan-Meier survival curves, "event-free (alive or without relapse)" means satisfying both of these conditions:
- Initially, the curve starts at 1.0 (100% survival) since all individuals are event-free at time zero.
- The steep decline in the early months indicates that a significant number of patients experience events, such as death or relapse, shortly after the transplant.
- This highlights the high-risk nature of the initial post-transplant period.
- As time progresses, the curve begins to level off, particularly after 20-30 months, suggesting that those who survive the initial phase tend to have better long-term outcomes.
- The survival probability never reaches zero, indicating that a portion of the population remains event-free throughout the observation period.
- The shaded region around the curve represents the confidence interval, which reflects the uncertainty of the survival estimates.
- Early on, the confidence intervals are narrow, indicating precise estimates due to a larger sample size.
- However, they widen at later time points, reflecting fewer patients being observed (due to censoring), which reduces the precision of the estimates.
- Overall, the Kaplan-Meier curve provides insight into the time-dependent risks of events, emphasizing the need for targeted interventions during the early post-transplant period to improve survival outcomes.
- The curve also suggests that patients who pass the high-risk early phase may achieve more favorable long-term survival.
- Further analysis, such as stratifying the data by race or comorbidity scores, could provide deeper insights into factors influencing survival and potential disparities across subgroups.
from lifelines import KaplanMeierFitter
# Instantiate the Kaplan-Meier fitter
kmf = KaplanMeierFitter()
# Kaplan-Meier fit for the entire dataset
plt.figure(figsize=(10, 6))
kmf.fit(durations=train['efs_time'], event_observed=train['efs'])
kmf.plot_survival_function()
plt.title('Kaplan-Meier Survival Curve for Entire Dataset')
plt.xlabel('Time (Months)')
plt.ylabel('Survival Probability')
plt.grid()
plt.show()
Kaplan-Meier Survival Curve: Stratified by Race
The Kaplan-Meier survival curve below visualizes the survival probabilities for different racial groups over time. Each line represents a specific race group. The shaded areas around the curves represent confidence intervals.
Key Observations:
- Early Survival Decline:
- All race groups show a steep initial decline in survival probability, indicating a high risk of adverse events shortly after transplantation.
- The rate of decline varies among groups, suggesting potential disparities in early survival outcomes.
- Group Differences in Long-Term Survival:
- Groups like "More than one race" and "Asian" exhibit higher long-term survival probabilities compared to "White" and "Black or African-American" groups.
- "American Indian or Alaska Native" and "Native Hawaiian or other Pacific Islander" groups show moderate survival probabilities.
- Confidence Intervals:
- Confidence intervals widen over time, reflecting reduced sample sizes.
- Widening is more pronounced in smaller racial groups, indicating greater uncertainty in survival estimates.
- Potential Disparities:
- The observed differences in survival probabilities suggest disparities in post-transplant outcomes that may be influenced by various factors.
- "White" and "Black or African-American" groups consistently have lower survival probabilities, highlighting areas for potential intervention.
# Kaplan-Meier fit for different groups (e.g., race_group)
plt.figure(figsize=(12, 8))
for group in train['race_group'].dropna().unique():
group_data = train[train['race_group'] == group]
kmf.fit(durations=group_data['efs_time'], event_observed=group_data['efs'], label=group)
kmf.plot_survival_function()
plt.title('Kaplan-Meier Survival Curve by Race Group')
plt.xlabel('Time (Months)')
plt.ylabel('Survival Probability')
plt.legend(title='Race Group')
plt.grid()
plt.show()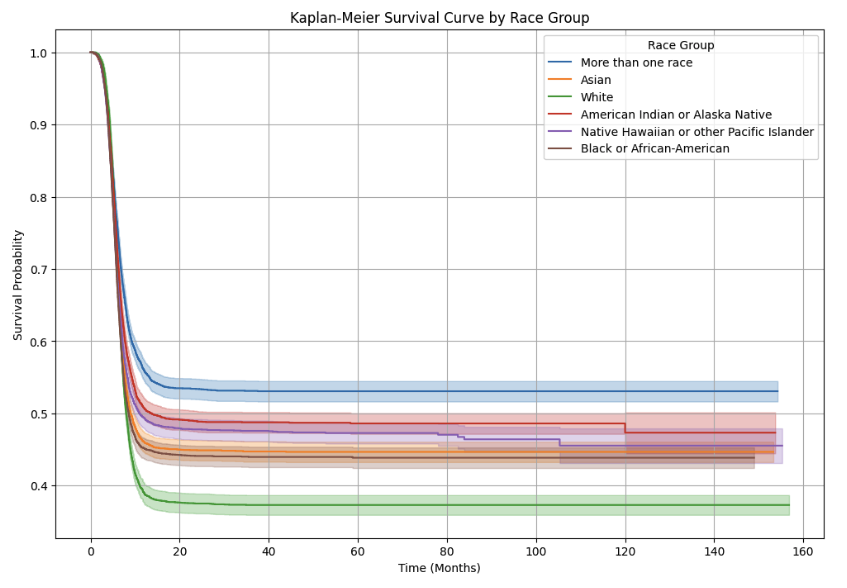
Kaplan-Meier Survival Curve: Stratified by Donor/Recipient Sex Match
The Kaplan-Meier survival curve below visualizes the survival probabilities for different donor/recipient sex match combinations over time. Each curve represents one of the four possible combinations:
- Male-to-Female (M-F)
- Female-to-Female (F-F)
- Female-to-Male (F-M)
- Male-to-Male (M-M)
The shaded areas around the curves indicate confidence intervals.
Key Observations:
- Early Decline in Survival:
- All groups show a steep initial decline in survival probability, reflecting the high-risk post-transplant period.
- Long-Term Survival Differences:
- F-F and M-M show the highest long-term survival probability.
- F-M and M-F have lower long-term survival probabilities.
- Confidence Intervals:
- Confidence intervals widen over time, particularly for M-F and F-M.
- F-F has relatively narrow intervals.
Sex Match Impact:
- F-F and M-M transplants tend to have better outcomes.
- M-F and F-M groups have lower survival probabilities.
Insights and Implications:
- Clinical Relevance:
- The survival advantage for F-F and M-M may reflect better immunological compatibility.
- M-F and F-M groups might benefit from additional clinical interventions.
- Meaning that M-F, F-M groups may require additional clinical interventions(treatments)
- Biological Factors:
- Differences in survival may stem from biological factors like immunological response or GVHD risk.
- immunological response:
- Refers to how our body's immune system responds to foreign substances
- In transplant situations:
- The immune reaction that occurs when donor cells enter the recipient's body
- If this response is too strong or too weak, it can negatively affect transplant outcomes
- GVHD (Graft Versus Host Disease) risk:
- A condition where transplanted donor immune cells recognize the recipient's body as 'foreign' and attack it
- Major symptoms:
- Skin rash
- Liver damage
- Digestive system problems
- A serious complication that can be life-threatening in severe cases
- immunological response:
- Differences in survival may stem from biological factors like immunological response or GVHD risk.
- Further Analysis:
- Additional factors should be analyzed alongside sex match.
- Statistical tests can confirm the significance of observed differences.
# Kaplan-Meier fit for a binary feature (e.g., gender)
plt.figure(figsize=(12, 8))
for gender in train['sex_match'].dropna().unique():
gender_data = train[train['sex_match'] == gender]
kmf.fit(durations=gender_data['efs_time'], event_observed=gender_data['efs'], label=gender)
kmf.plot_survival_function()
plt.title('Kaplan-Meier Survival Curve by Sex Match')
plt.xlabel('Time (Months)')
plt.ylabel('Survival Probability')
plt.legend(title='Sex Match')
plt.grid()
plt.show()
Cox Proportional Hazards (CPH) Model
- The Cox Proportional Hazards (CPH) model is a widely used method in survival analysis for evaluating the effect of multiple covariates on the time to a specific event, such as death or relapse.
- Unlike non-parametric methods like Kaplan-Meier, CPH is a semi-parametric model incorporating covariates to estimate their influence on survival while making no assumptions about the baseline hazard function.
- baseline hazard function: the change in basic risk rate over time



Hazard Function h(t|X) in detail:
- Basic Concept:
- Represents the instantaneous probability that someone who has survived until time t will experience an event right after
- Here, 'event' could be death, disease recurrence, etc.
- Formula Components:
- h₀(t): baseline hazard function
- Basic risk rate when all covariates are 0
- Can change over time
- exp(β₁X₁ + β₂X₂ + ... + βₚXₚ): effect of covariates
- X₁, X₂, ..., Xₚ: covariates (age, gender, etc.)
- β₁, β₂, ..., βₚ: coefficients showing the influence of each covariate
- Why use exp: ensures hazard rate is always positive
- h₀(t): baseline hazard function
- Practical Meaning:
- For example, in hematopoietic cell transplant patients:
- h(t): risk of death/relapse at time t
- X₁: patient's age
- X₂: gender matching status
- β₁: impact of age on risk
- β₂: impact of gender matching on risk
- For example, in hematopoietic cell transplant patients:
Proportional Hazards Assumption in detail:
- Assumes that the hazard ratio between two patients remains constant over time
- Example:
- If a 50-year-old patient has twice the risk of a 30-year-old patient
- This "twice" ratio remains constant whether it's 1 month or 1 year post-transplant
- Therefore "TIME-INDEPENDENT"
Hazard Ratio (HR) in detail:
- Calculated as HR = exp(β)
- HR > 1: increased risk
- Example: HR = 2 means double the risk
- HR < 1: decreased risk
- Example: HR = 0.5 means half the risk
- HR = 1: no effect
- If β = 0.693 for gender matching:
- HR = exp(0.693) = 2
- This means for gender mismatch:
- Risk doubles
- This doubling remains constant at any time post-transplant
Censoring in detail:
What is censoring?
- When the event of interest (e.g., death, relapse) doesn't occur during the study period
- In other words, when we can't know the patient's final outcome
Cases of right-censoring:
- No event occurs until the end of the study
- Example: Patient survives throughout a 5-year follow-up study
- Patient drops out during follow-up
- Example: Transfer to another hospital
- Example: Loss of contact
- Excluded from study for other reasons
- Example: Patient requests to discontinue participation
Handling in Cox model:
- Censored data is included in the analysis
- Information up to the censoring point is used for model estimation
- Unbiased estimates are calculated through likelihood function
Likelihood function in detail:
What is a likelihood function:
- A function that calculates the possibility (probability) that observed data came from a specific statistical model
- In other words, it quantifies "how likely this data would come from this model"
- Let's assume we have patient survival data:
- Patient A: Died after 2 years
- Patient B: Survived until 3 years (then lost to follow-up)
- Patient C: Died after 5 years
The likelihood function:
1. Calculates the probability of each patient's observed outcome
2. Multiplies all these probabilities
3. The higher this value, the better the model explains the data
In Cox model:
- Censored data (e.g., Patient B) is included in the likelihood function
- Uses information up to the point of censoring
- This enables unbiased parameter estimation
In this way, the likelihood function allows us to effectively use incomplete data (censored data) in the analysis.
Partial Likelihood in detail:
- Considers only the order of event occurrences instead of complete time information
- In other words, focuses more on "who experienced the event first" rather than "exact timing"
- Assume we have three patients:
Patient A: Dies at 2 months
Patient B: Dies at 5 months
Patient C: Survives until 7 months (censored)
Partial likelihood analyzes:
- At 2 months: "Why did A die instead of the others"
- At 5 months: "Why did B die among remaining patients" - Reasons for This Approach:
- No need to specify baseline hazard function (h₀(t))
- Can estimate covariate effects (β) using just event order
- Simpler and more efficient computation
- Maximization Process:
- Find β values that maximize the partial likelihood
- These β values are considered to best explain each variable's effect on survival
from lifelines import CoxPHFitter
# Preprocess data
# Select relevant columns for Cox regression
cox_features = ['efs_time', 'efs', 'age_at_hct', 'karnofsky_score', 'comorbidity_score', 'race_group']
train = train[cox_features]
# Convert categorical variables into dummy variables
train = pd.get_dummies(train, columns=['race_group'], drop_first=True)
# Drop rows with missing values (ensure clean data for Cox model)
train = train.dropna()
# Instantiate and fit the Cox Proportional Hazards model
cph = CoxPHFitter()
cph.fit(train, duration_col='efs_time', event_col='efs')
# Show summary of the model
cph.print_summary()
Resultsr
- The hazard ratio (HR) plot illustrates the effects of different covariates on the hazard of the event occurring, as estimated by the Cox Proportional Hazards model.
- The x-axis represents the hazard ratio, where a value of 1.0 (marked by the dashed vertical line) indicates no effect on the hazard.
- Hazard ratios greater than 1.0 indicate an increased risk of the event, while values less than 1.0 suggest a protective effect or reduced risk.
- The 95% confidence intervals (CIs) are shown as horizontal lines around each hazard ratio, indicating the uncertainty in the estimates.
- If a confidence interval crosses 1.0, the effect of the covariate is not statistically significant.
- The analysis reveals several key findings.
- Among race groups, "Black or African-American" and "White" have hazard ratios slightly above 1.0, indicating a marginally increased risk compared to the reference group (likely another race, such as "Asian" or "More than one race").
- Conversely, the "More than one race" group has an HR less than 1.0, suggesting a protective effect, while "Native Hawaiian or other Pacific Islander" shows little to no impact on the hazard.
- The comorbidity score has an HR slightly above 1.0, indicating that patients with more comorbidities are at greater risk of the event.
- Comorbidity: A condition where (a patient) suffers from two chronic diseases simultaneously
- Similarly, "age at HCT" has a hazard ratio above 1.0, suggesting that older patients face a slightly higher risk.
- In contrast, the Karnofsky performance score has a hazard ratio less than 1.0, reflecting a protective effect where higher scores (indicating better performance status) are associated with reduced risk.
- The Karnofsky performance score (KPS) or Karnofsky performance status scale is a measure to evaluate a patient's overall functional status.
- Scoring System (0-100):
- 100: Normal, no symptoms or signs of disease
- 90: Able to carry on normal activity, minor symptoms/signs
- 80: Normal activity with effort, some symptoms/signs
- 70: Cares for self but unable to carry on normal activity or work
- 60: Requires occasional assistance but can meet most personal needs
- 50: Requires considerable assistance and frequent medical care
- 40: Disabled, requires special care and assistance
- 30: Severely disabled, hospital admission indicated, death not imminent
- 20: Very sick, hospital admission necessary, active supportive treatment needed
- 10: Moribund, death imminent
- 0: Dead
- 100: Normal, no symptoms or signs of disease
- Scoring System (0-100):
- The Karnofsky performance score (KPS) or Karnofsky performance status scale is a measure to evaluate a patient's overall functional status.
- Statistical significance can be inferred from the confidence intervals.
- Covariates such as comorbidity score and Karnofsky score likely have statistically significant effects, as their confidence intervals do not cross 1.0.
- Some race groups and "age at HCT", however, may not have significant effects, as their intervals overlap with 1.0.
- These findings suggest that clinical factors, particularly comorbidity score and performance status, are key predictors of survival outcomes.
- Additionally, differences in hazard ratios among race groups point to potential disparities in outcomes that warrant further investigation.
- Efforts to reduce comorbidities, improve performance status, and explore the underlying causes of racial disparities could help optimize patient care and outcomes.
- This analysis highlights the importance of targeted interventions and provides a foundation for further exploration of survival determinants.
# Visualize the coefficients (hazard ratios)
cph.plot(hazard_ratios=True)
plt.title("Cox Regression - Hazard Ratios")
plt.show()
Survival Curves for Comorbidity Score
- The survival curves generated by the Cox Proportional Hazards (CPH) model illustrate the relationship between comorbidity score and survival probabilities over time.
- The x-axis represents time (e.g., in months), while the y-axis shows the probability of survival.
- Each line corresponds to a specific comorbidity score, ranging from 0 (no comorbidities) to 4 (high comorbidity burden), with a dashed line representing the baseline survival curve.
- The results indicate that higher comorbidity scores are associated with lower survival probabilities, as reflected by the descending order of the survival curves.
- Patients with a comorbidity score of 0 exhibit the highest survival probabilities, while those with a score of 4 experience the steepest decline and the lowest overall survival.
- All survival curves show a steep decline during the early months, reflecting a high-risk period immediately after the transplant.
- This decline is more pronounced for patients with higher comorbidity scores, indicating that comorbidities significantly exacerbate early post-transplant risks.
- Beyond the initial phase, the survival curves stabilize, but patients with higher comorbidity scores continue to have significantly lower survival probabilities compared to those with lower scores.
- The persistent gap between the survival curves suggests that comorbidities have a lasting impact on survival outcomes.
- The baseline survival curve aligns closely with a mid-range comorbidity score, representing an "average" patient in the population.
- These findings highlight the clinical importance of managing comorbidities before and after transplantation.
- Higher comorbidity scores predict worse survival outcomes, emphasizing the need for targeted interventions and closer monitoring for high-risk patients, particularly during the early post-transplant phase.
- Even long-term outcomes are worse for patients with higher scores, indicating the necessity of sustained care.
- This analysis also underscores the potential for risk stratification, where patients can be categorized by comorbidity scores to prioritize resources and tailor interventions.
cph.plot_partial_effects_on_outcome(covariates='comorbidity_score', values=[0, 1, 2, 3, 4], cmap='coolwarm');
탁월성은 평범함에서 나온다
<GRIT>