First competition following Yuhan Lee's kaggle curriculum. Binary classification competition using tabular data.
First Kernel
- EDA(Exploratory Data Analysis) written on Titanic competition by Yuhan himself.
- visualization tool used: matplotlib, seaborn, plotly
- data analysis tool used: pandas, numpy
- machine learning tool: sklearn
- code: https://github.com/LDYWINNER/TIML/blob/main/kaggle-roadmap/binary-classification/titanic/eda%20tutorial.ipynb
Insights / Summary:
1.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use('seaborn')
sns.set(font_scale=2.5)-> base configuration above all that sets seaborn scheme instead of matplotlib's default scheme and base font scale.
2.
- pandas is the most used library that works well with tabular data
- describe() function which can be used with pandas' dataframe returns statistics of each feature of the dataset
3.
for col in df_train.columns:
msg = 'column: {:>10}\t Percent of NaN value: {:.2f}%'.format(col, 100 * (df_train[col].isnull().sum() / df_train[col].shape[0]))
print(msg)-> null data checking code for train dataset
4.
import missingno as msno
msno.matrix(df=df_train.iloc[:, :], figsize=(8, 8), color=(0.8, 0.5, 0.2))
msno.bar(df=df_train.iloc[:, :], figsize=(8, 8), color=(0.8, 0.5, 0.2))-> helps us to check null data easier
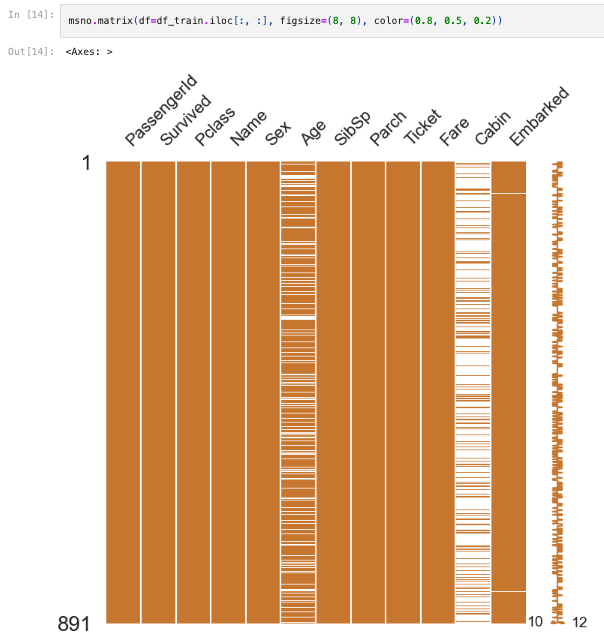

5.
- We should check whether the distribution of target label from test dataset is balanced.
- It critically affects the evaluation of the model.
- For example, if the distribution of target label of the test dataset is 99% 1 and the model returns 1 no matter what the input value is, the model's accuracy is 99%.
6.
- Ordinal data(서수형 데이터) is a type of categorical data where the values follow a natural order or ranking, but the differences between values aren't necessarily equal.
7.
df_train[['Pclass', 'Survived']].groupby(['Pclass'], as_index=True).count()
# easier way - to use crosstab
pd.crosstab(df_train['Pclass'], df_train['Survived'], margins=True).style.background_gradient(cmap='summer_r')-> counting values for a specific feature(categorical variable)
8.
df_train[['Pclass', 'Survived']].groupby(['Pclass'], as_index=True).mean().sort_values(by='Survived', ascending=False).plot.bar()-> rate of survival
9.
seaborn's countplot is a good way to check the number of each label of specific feature.
y_position = 1.02
f, ax = plt.subplots(1, 2, figsize=(18, 8))
df_train['Pclass'].value_counts().plot.bar(color=['#CD7F32','#FFDF00','#D3D3D3'], ax=ax[0])
ax[0].set_title('Number of Passengers By Pclass', y=y_position)
ax[0].set_ylabel('Count')
sns.countplot('Pclass', hue='Survived', data=df_train, ax=ax[1])
ax[1].set_title('Pclass: Survived vs Dead', y=y_position)
plt.show()
10.
seaborn's factorplot is a good way to plot graphs with 3 dimensions.
sns.factorplot('Pclass', 'Survived', hue='Sex', data=df_train, size=6, aspect=1.5)
violinplot can be an alternative: by using violinplot, we can divide the case for each label(x axis) as well as see the distribution of another feature(y axis)
f, ax = plt.subplots(1, 2, figsize=(18, 8))
sns.violinplot(x="Pclass", y="Age", hue="Survived", data=df_train, scale='count', split=True, ax=ax[0])
ax[0].set_title('Pclass and Age vs. Survived')
ax[0].set_yticks(range(0, 110, 10))
sns.violinplot(x="Sex", y="Age", hue="Survived", data=df_train, scale='count', split=True, ax=ax[1])
ax[1].set_title('Sex and Age vs. Survived')
ax[1].set_yticks(range(0, 110, 10))
plt.show()
11.
If we are looking at a feature that has continuous value, we can plot it to a graph and check the skewness.
fig, ax = plt.subplots(1, 1, figsize=(8, 8))
g = sns.distplot(df_train['Fare'], color='b', label='Skewness: {:.2f}'.format(df_train['Fare'].skew()), ax=ax)
g = g.legend(loc='best')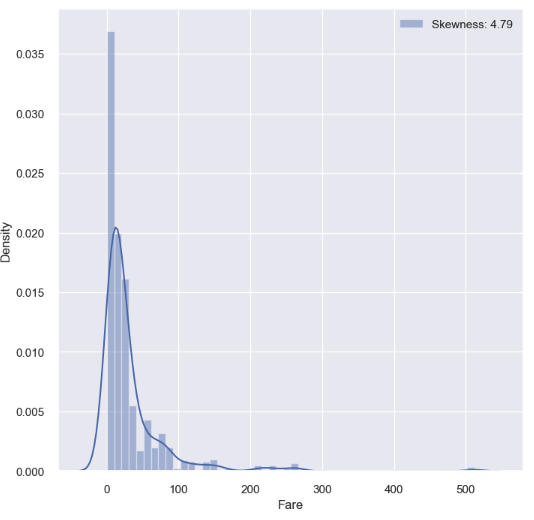
- This graph shows considerably high skewness.
- If we use features that have high skewness in the prediction model, the model can be excessively sensitive to outlier which will result in bad performance.
- In this case, we can apply log to all value of this feature: we can use map and apply methods from pandas
df_train['Fare'] = df_train['Fare'].map(lambda i: np.log(i) if i > 0 else 0)
df_test['Fare'] = df_test['Fare'].map(lambda i: np.log(i) if i > 0 else 0)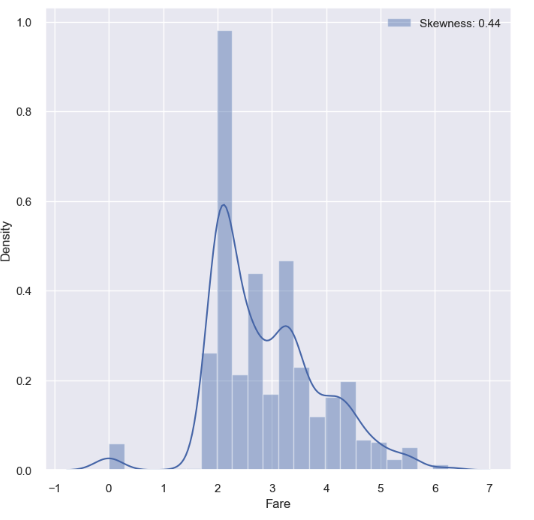
12.
If specific feature have excessively high proportion of null value, it can be better to just get rid of that feature while training.
Second Kernel
- gives idea how the workflow is in any predictive modeling problem
- code: https://github.com/LDYWINNER/TIML/blob/main/kaggle-roadmap/binary-classification/titanic/eda%20to%20prediction.ipynb
Insights:
1.
data.isnull().sum()-> maybe simpler way for checking null values throughout the dataset: only if the null value is stored as a null value(null value can be stored in various ways: -1, nan, ...)
2.
Types of features:
- Categorical features:
- one that has two or more categories and each value in that feature can be categorized by them
- ex) sex
- Ordinal features:
- similar to categorical values, but the difference between them is that we can have relative ordering or sorting between the values
- ex: height divided into 3 categories: "Tall", "Medium", "Short"
- Continuous feature
- a feature is said to be continuous if it can take values between any two points or betwen the minimum or maximum values in the features column
- ex: age
3.
- In this dataset, there are 177 null values in Age feature.
- We can simply assign total mean value of Age feature to those null values, but simply assigning mean value doesn't seem to be that legitimate.
- In this case, we can get help from other features.
- We have Name feature which includes salutations such as Mr. and Mrs that we can get hint of the age of the person.
- Thus, it is important to fully analyze the dataset before performing any null value imputations.
data['Initial']=0
for i in data:
data['Initial']=data.Name.str.extract('([A-Za-z]+)\.')
4.
data['Embarked'].fillna('S',inplace=True)- We can fill the null values with the mode(최빈값) in some cases.
- As we saw that maximum passengers boarded from Port S, we replace NaN with S.
5.
sns.heatmap(data.corr(),annot=True,cmap='RdYlGn',linewidths=0.2) #data.corr()-->correlation matrix
fig=plt.gcf()
fig.set_size_inches(10,8)
plt.show()- Taking a look at seaborn's heatmap is a good way to check the correlation between features.
- POSITIVE CORRELATION: If an increase in feature A leads to increase in feature B, then they are positively correlated. A value 1 means perfect positive correlation.
- NEGATIVE CORRELATION: If an increase in feature A leads to decrease in feature B, then they are negatively correlated. A value -1 means perfect negative correlation.
- Now lets say that two features are highly or perfectly correlated, so the increase in one leads to increase in the other. This means that both the features are containing highly similar information and there is very little or no variance in information. This is known as MultiColinearity as both of them contains almost the same information.
- So do you think we should use both of them as one of them is redundant. While making or training models, we should try to eliminate redundant features as it reduces training time and many such advantages.
6.
To use continuous feature in machine learning model, we should convert into categorical feature by binning or normalization.
Binning example:
data['Age_band']=0
data.loc[data['Age']<=16,'Age_band']=0
data.loc[(data['Age']>16)&(data['Age']<=32),'Age_band']=1
data.loc[(data['Age']>32)&(data['Age']<=48),'Age_band']=2
data.loc[(data['Age']>48)&(data['Age']<=64),'Age_band']=3
data.loc[data['Age']>64,'Age_band']=4
When making into ordinal feature instead of categorical feature, we can use pandas library's qcut method: splits or arranges the values according the number of bins we have passed.
Binning example:
data['Fare_Range']=pd.qcut(data['Fare'],4)
data.groupby(['Fare_Range'])['Survived'].mean().to_frame().style.background_gradient(cmap='summer_r')
data['Fare_cat']=0
data.loc[data['Fare']<=7.91,'Fare_cat']=0
data.loc[(data['Fare']>7.91)&(data['Fare']<=14.454),'Fare_cat']=1
data.loc[(data['Fare']>14.454)&(data['Fare']<=31),'Fare_cat']=2
data.loc[(data['Fare']>31)&(data['Fare']<=513),'Fare_cat']=3
7.
When using KNN model, we can check if which value of k will work well.
Code example:
a_index=list(range(1,11))
a=pd.Series()
x=[0,1,2,3,4,5,6,7,8,9,10]
for i in list(range(1,11)):
model=KNeighborsClassifier(n_neighbors=i)
model.fit(train_X,train_Y)
prediction=model.predict(test_X)
a=a.append(pd.Series(metrics.accuracy_score(prediction,test_Y)))
plt.plot(a_index, a)
plt.xticks(x)
fig=plt.gcf()
fig.set_size_inches(12,6)
plt.show()
print('Accuracies for different values of n are:',a.values,'with the max value as ',a.values.max())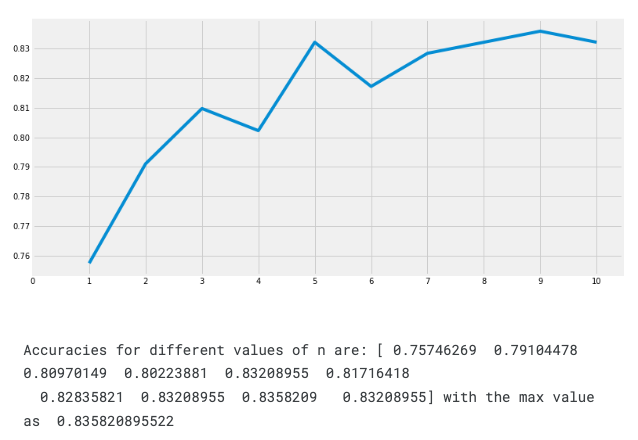
8.
- Accuracy of classification prediction can be sometimes misleading due to imbalance.
- We can get a summarized result with confusion matrix, which shows where did the model go wrong or which class did the model predict wrong.
- Gives the number of correct and incorrect classifications made by the classifier.
f,ax=plt.subplots(3,3,figsize=(12,10))
y_pred = cross_val_predict(svm.SVC(kernel='rbf'),X,Y,cv=10)
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[0,0],annot=True,fmt='2.0f')
ax[0,0].set_title('Matrix for rbf-SVM')
y_pred = cross_val_predict(svm.SVC(kernel='linear'),X,Y,cv=10)
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[0,1],annot=True,fmt='2.0f')
ax[0,1].set_title('Matrix for Linear-SVM')
y_pred = cross_val_predict(KNeighborsClassifier(n_neighbors=9),X,Y,cv=10)
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[0,2],annot=True,fmt='2.0f')
ax[0,2].set_title('Matrix for KNN')
y_pred = cross_val_predict(RandomForestClassifier(n_estimators=100),X,Y,cv=10)
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[1,0],annot=True,fmt='2.0f')
ax[1,0].set_title('Matrix for Random-Forests')
y_pred = cross_val_predict(LogisticRegression(),X,Y,cv=10)
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[1,1],annot=True,fmt='2.0f')
ax[1,1].set_title('Matrix for Logistic Regression')
y_pred = cross_val_predict(DecisionTreeClassifier(),X,Y,cv=10)
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[1,2],annot=True,fmt='2.0f')
ax[1,2].set_title('Matrix for Decision Tree')
y_pred = cross_val_predict(GaussianNB(),X,Y,cv=10)
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[2,0],annot=True,fmt='2.0f')
ax[2,0].set_title('Matrix for Naive Bayes')
plt.subplots_adjust(hspace=0.2,wspace=0.2)
plt.show()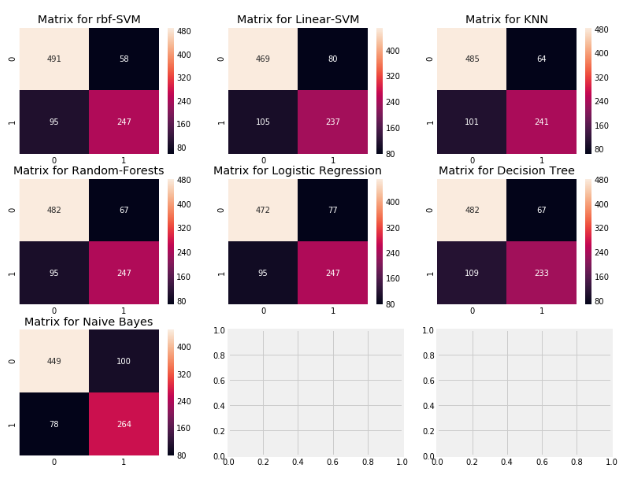
- Interpretation:
- The left diagonal shows the number of correct predictions made for each class while the right diagonal shows the number of wrong prredictions made. Lets consider the first plot for rbf-SVM:
- 1)The no. of correct predictions are 491(for dead) + 247(for survived) with the mean CV accuracy being (491+247)/891 = 82.8% which we did get earlier.
- 2)Errors--> Wrongly Classified 58 dead people as survived and 95 survived as dead. Thus it has made more mistakes by predicting dead as survived.
- By looking at all the matrices, we can say that rbf-SVM has a higher chance in correctly predicting dead passengers but NaiveBayes has a higher chance in correctly predicting passengers who survived.
9.
VotingClassifier
- Simplest way of combining predictions from simple ml models
- Gives average prediction result based on the prediction of all submodels
- Code example:
from sklearn.ensemble import VotingClassifier
ensemble_lin_rbf=VotingClassifier(estimators=[('KNN',KNeighborsClassifier(n_neighbors=10)),
('RBF',svm.SVC(probability=True,kernel='rbf',C=0.5,gamma=0.1)),
('RFor',RandomForestClassifier(n_estimators=500,random_state=0)),
('LR',LogisticRegression(C=0.05)),
('DT',DecisionTreeClassifier(random_state=0)),
('NB',GaussianNB()),
('svm',svm.SVC(kernel='linear',probability=True))
],
voting='soft').fit(train_X,train_Y)
print('The accuracy for ensembled model is:',ensemble_lin_rbf.score(test_X,test_Y))
cross=cross_val_score(ensemble_lin_rbf,X,Y, cv = 10,scoring = "accuracy")
print('The cross validated score is',cross.mean())
Bagging
- Works by applying similar classifiers on small partitions of the dataset and then taking average of all the predictions
- Due to the averaging, there is a reduction in variance
- Unlike Voting Classifier, Bagging makes use of similar classifers
- Bagging works best with models with high variance: maybe decision tree or random forests
Decision tree code example:
from sklearn.ensemble import BaggingClassifier
model=BaggingClassifier(base_estimator=KNeighborsClassifier(n_neighbors=3),random_state=0,n_estimators=700)
model.fit(train_X,train_Y)
prediction=model.predict(test_X)
print('The accuracy for bagged KNN is:',metrics.accuracy_score(prediction,test_Y))
result=cross_val_score(model,X,Y,cv=10,scoring='accuracy')
print('The cross validated score for bagged KNN is:',result.mean())
Boosting
- Uses sequential learning of classifiers
- step by step enhancement of a weak model
- Steps:
- A model is first trained on the complete dataset.
- Now the model will get some instances right while some wrong.
- Now in the next iteration, the learner will focus more on the wrongly predicted instances or give more weight to it.
- Thus it will try to predict the wrong instance correctly.
- Now this iterative process continuous, and new classifers are added to the model until the limit is reached on the accuracy.
10.
Feature Importance code example:
f,ax=plt.subplots(2,2,figsize=(15,12))
model=RandomForestClassifier(n_estimators=500,random_state=0)
model.fit(X,Y)
pd.Series(model.feature_importances_,X.columns).sort_values(ascending=True).plot.barh(width=0.8,ax=ax[0,0])
ax[0,0].set_title('Feature Importance in Random Forests')
model=AdaBoostClassifier(n_estimators=200,learning_rate=0.05,random_state=0)
model.fit(X,Y)
pd.Series(model.feature_importances_,X.columns).sort_values(ascending=True).plot.barh(width=0.8,ax=ax[0,1],color='#ddff11')
ax[0,1].set_title('Feature Importance in AdaBoost')
model=GradientBoostingClassifier(n_estimators=500,learning_rate=0.1,random_state=0)
model.fit(X,Y)
pd.Series(model.feature_importances_,X.columns).sort_values(ascending=True).plot.barh(width=0.8,ax=ax[1,0],cmap='RdYlGn_r')
ax[1,0].set_title('Feature Importance in Gradient Boosting')
model=xg.XGBClassifier(n_estimators=900,learning_rate=0.1)
model.fit(X,Y)
pd.Series(model.feature_importances_,X.columns).sort_values(ascending=True).plot.barh(width=0.8,ax=ax[1,1],color='#FD0F00')
ax[1,1].set_title('Feature Importance in XgBoost')
plt.show()
Third Kernel
- Titanic top 4% solution with ensemble modeling
- Code: https://github.com/LDYWINNER/TIML/blob/main/kaggle-roadmap/binary-classification/titanic/Titanic%20top%204%25%20with%20ensemble%20modeling.ipynb
Insights:
1.
Outlier detection code example:
# Outlier detection
def detect_outliers(df,n,features):
"""
Takes a dataframe df of features and returns a list of the indices
corresponding to the observations containing more than n outliers according
to the Tukey method.
"""
outlier_indices = []
# iterate over features(columns)
for col in features:
# 1st quartile (25%)
Q1 = np.percentile(df[col], 25)
# 3rd quartile (75%)
Q3 = np.percentile(df[col],75)
# Interquartile range (IQR)
IQR = Q3 - Q1
# outlier step
outlier_step = 1.5 * IQR
# Determine a list of indices of outliers for feature col
outlier_list_col = df[(df[col] < Q1 - outlier_step) | (df[col] > Q3 + outlier_step )].index
# append the found outlier indices for col to the list of outlier indices
outlier_indices.extend(outlier_list_col)
# select observations containing more than 2 outliers
outlier_indices = Counter(outlier_indices)
multiple_outliers = list( k for k, v in outlier_indices.items() if v > n )
return multiple_outliers
# detect outliers from Age, SibSp , Parch and Fare
Outliers_to_drop = detect_outliers(train,2,["Age","SibSp","Parch","Fare"])- This kernel detected rows that have more than 2 outliers and decided to drop them.
- For the case of "Fare" feature, it had 1 nan value in the whole dataset: we simply filled it with median value in this case.
- For the case of "Embarked" feature, it had 2 nan value in the whole dataset: we filled with most frequent value "S".
- For the case of "Age" feature, it had 256 nan values in the whole dataset. So, we need a legitimate way to impute the missing values.
- First we checked the most correlated features with Age: Sex, Parch, Pclass, SibSp.
- Using seaborn heatmap, we found out Sex feature had no correlation with Age feature and had negative correlation with Pclass, Parch, and SibSp features.
- When we use factorplot to take a look at correlation between Age feature and those 3 features, we can see positive correlation but heatmap shows negative correlation.
- This can be caused due to 2 reasons: 1. The distribution of data may be imbalanced 2. It could be a non-linear relationship.
- So we decided to use SibSp, Parch, and Pclass to impute the missing Age values: Fill age with the median age of similar rows according to Pclass, Parch, and SibSp.
Code Example:
# Filling missing value of Age
## Fill Age with the median age of similar rows according to Pclass, Parch and SibSp
# Index of NaN age rows
index_NaN_age = list(dataset["Age"][dataset["Age"].isnull()].index)
for i in index_NaN_age :
age_med = dataset["Age"].median()
age_pred = dataset["Age"][((dataset['SibSp'] == dataset.iloc[i]["SibSp"]) & (dataset['Parch'] == dataset.iloc[i]["Parch"]) & (dataset['Pclass'] == dataset.iloc[i]["Pclass"]))].median()
if not np.isnan(age_pred) :
dataset['Age'].iloc[i] = age_pred
else :
dataset['Age'].iloc[i] = age_med
2.
Checking for null and missing values: this kernel first filled empty and nan values with np.nan to prevent missing them.
# Fill empty and NaNs values with NaN
dataset = dataset.fillna(np.nan)
# Check for Null values
dataset.isnull().sum()
3.
Code example of converting categorical variables into indicator values
# convert to indicator values Title and Embarked
dataset = pd.get_dummies(dataset, columns = ["Title"])
dataset = pd.get_dummies(dataset, columns = ["Embarked"], prefix="Em")
4.
How this kernel made prediction model:
- Compared 10 popular classifiers and evaluate the mean accuracy of each of them by stratified kfold cross validation.
- SVC
- Decision Tree
- AdaBoost
- RandomForest
- Extra Trees
- Gradient Boosting
- Multiple Layer Perceptron(Neural Network)
- KNN
- Logistic Regression
- Linear Discriminant Analysis
- Decided best 5: SVC, AdaBoost, RandomForest, ExtraTrees and GradientBoosting for ensemble modeling.
- Hyperparameter tunning for those 5 models: grid search optimization
- Plot learning curves: learning curves are a good way to see the overfitting effect on the training set and the effect of the training size on the accuracy.
- Took a look at feature importance of the classifiers.
- Combined model with VotingClassifier: used soft voting.
Grid Search Optimization Code Example of AdaBoost:
### META MODELING WITH ADABOOST, RF, EXTRATREES and GRADIENTBOOSTING
# Adaboost
DTC = DecisionTreeClassifier()
adaDTC = AdaBoostClassifier(DTC, random_state=7)
ada_param_grid = {"base_estimator__criterion" : ["gini", "entropy"],
"base_estimator__splitter" : ["best", "random"],
"algorithm" : ["SAMME","SAMME.R"],
"n_estimators" :[1,2],
"learning_rate": [0.0001, 0.001, 0.01, 0.1, 0.2, 0.3,1.5]}
gsadaDTC = GridSearchCV(adaDTC,param_grid = ada_param_grid, cv=kfold, scoring="accuracy", n_jobs= 4, verbose = 1)
gsadaDTC.fit(X_train,Y_train)
ada_best = gsadaDTC.best_estimator_
Plotting Learning Curves Code Example:
def plot_learning_curve(estimator, title, X, y, ylim=None, cv=None,
n_jobs=-1, train_sizes=np.linspace(.1, 1.0, 5)):
"""Generate a simple plot of the test and training learning curve"""
plt.figure()
plt.title(title)
if ylim is not None:
plt.ylim(*ylim)
plt.xlabel("Training examples")
plt.ylabel("Score")
train_sizes, train_scores, test_scores = learning_curve(
estimator, X, y, cv=cv, n_jobs=n_jobs, train_sizes=train_sizes)
train_scores_mean = np.mean(train_scores, axis=1)
train_scores_std = np.std(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
plt.grid()
plt.fill_between(train_sizes, train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std, alpha=0.1,
color="r")
plt.fill_between(train_sizes, test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std, alpha=0.1, color="g")
plt.plot(train_sizes, train_scores_mean, 'o-', color="r",
label="Training score")
plt.plot(train_sizes, test_scores_mean, 'o-', color="g",
label="Cross-validation score")
plt.legend(loc="best")
return plt
g = plot_learning_curve(gsRFC.best_estimator_,"RF mearning curves",X_train,Y_train,cv=kfold)
g = plot_learning_curve(gsExtC.best_estimator_,"ExtraTrees learning curves",X_train,Y_train,cv=kfold)
g = plot_learning_curve(gsSVMC.best_estimator_,"SVC learning curves",X_train,Y_train,cv=kfold)
g = plot_learning_curve(gsadaDTC.best_estimator_,"AdaBoost learning curves",X_train,Y_train,cv=kfold)
g = plot_learning_curve(gsGBC.best_estimator_,"GradientBoosting learning curves",X_train,Y_train,cv=kfold)
Fourth Kernel
- About ensembling and stacking
- Code: https://github.com/LDYWINNER/TIML/blob/main/kaggle-roadmap/binary-classification/titanic/Intro%20to%20ensembling%20and%20stacking.ipynb
Insights:
1.
- Stacking uses predictions of base classifiers a input for training to a second-level model.
- However, one cannot simply train the base models on the full training data, generate predictions on the full test set and then output these for the second-level training.
- This runs the risk of the base model predictions already having "seen" the test set and therefore overfitting when feeding these predictions.
- Out-of-fold predictions: refers to the predicted values obtained during the cross-validation process
def get_oof(clf, x_train, y_train, x_test):
oof_train = np.zeros((ntrain,))
oof_test = np.zeros((ntest,))
oof_test_skf = np.empty((NFOLDS, ntest))
for i, (train_index, test_index) in enumerate(kf):
x_tr = x_train[train_index]
y_tr = y_train[train_index]
x_te = x_train[test_index]
clf.train(x_tr, y_tr)
oof_train[test_index] = clf.predict(x_te)
oof_test_skf[i, :] = clf.predict(x_test)
oof_test[:] = oof_test_skf.mean(axis=0)
return oof_train.reshape(-1, 1), oof_test.reshape(-1, 1)- We define a method that records the out-of-fold predictions.
- Then, we use the out-of-fold predictions to train the stacking model.
base_predictions_train = pd.DataFrame( {'RandomForest': rf_oof_train.ravel(),
'ExtraTrees': et_oof_train.ravel(),
'AdaBoost': ada_oof_train.ravel(),
'GradientBoost': gb_oof_train.ravel()
})- Concatenate and join both first-level train and test predictions as x_train and x_test.
- Then, fit a second-level training model: used XGBoost, boosted tree learning model. It is built to optimize large-scale boosted tree algorithms.
- The more uncorrelated the results of first-level models, the better the final score will be.
x_train = np.concatenate(( et_oof_train, rf_oof_train, ada_oof_train, gb_oof_train, svc_oof_train), axis=1)
x_test = np.concatenate(( et_oof_test, rf_oof_test, ada_oof_test, gb_oof_test, svc_oof_test), axis=1)gbm = xgb.XGBClassifier(
#learning_rate = 0.02,
n_estimators= 2000,
max_depth= 4,
min_child_weight= 2,
#gamma=1,
gamma=0.9,
subsample=0.8,
colsample_bytree=0.8,
objective= 'binary:logistic',
nthread= -1,
scale_pos_weight=1).fit(x_train, y_train)
predictions = gbm.predict(x_test)
No pressure, no diamonds.
- Thomas Carlyle -
'캐글' 카테고리의 다른 글
| [Kaggle Study] 3. Learning Rate (2) | 2024.10.29 |
|---|---|
| [Kaggle Study] 2. Scale of Features (1) | 2024.10.29 |
| [Kaggle Study] 1. Loss Function 손실 함수 & Gradient Descent 경사 하강법 (6) | 2024.10.25 |
| ML/DL(3) - 손실 함수와 경사 하강법의 관계 (0) | 2023.07.08 |
| ML/DL(2) - 오차 역전파(backpropagation) (0) | 2023.07.08 |